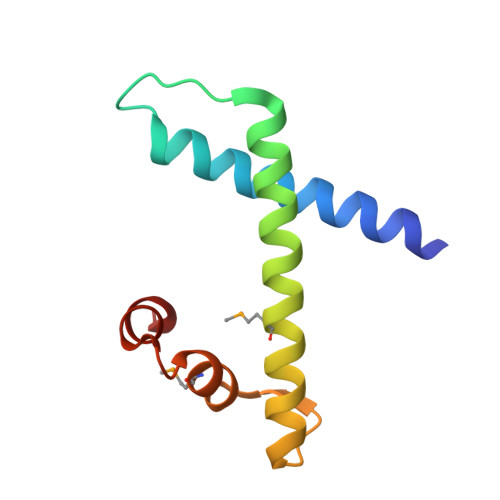
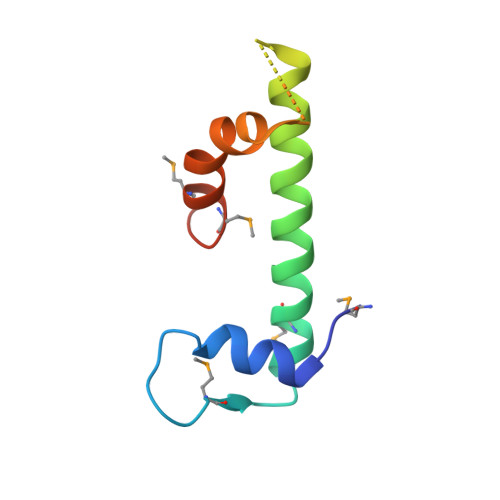
Saccharomyces Cerevisiae MHF Complex Structurally Resembles the Histones (H3-H4)(2) Heterotetramer and Functions as a Heterotetramer
Yang, H., Zhang, T., Tao, Y., Wu, L., Li, H.T., Zhou, J.Q., Zhong, C., Ding, J.(2012) Structure 20: 364-370
- PubMed: 22325783
- DOI: https://doi.org/10.1016/j.str.2011.12.012
- Primary Citation of Related Structures:
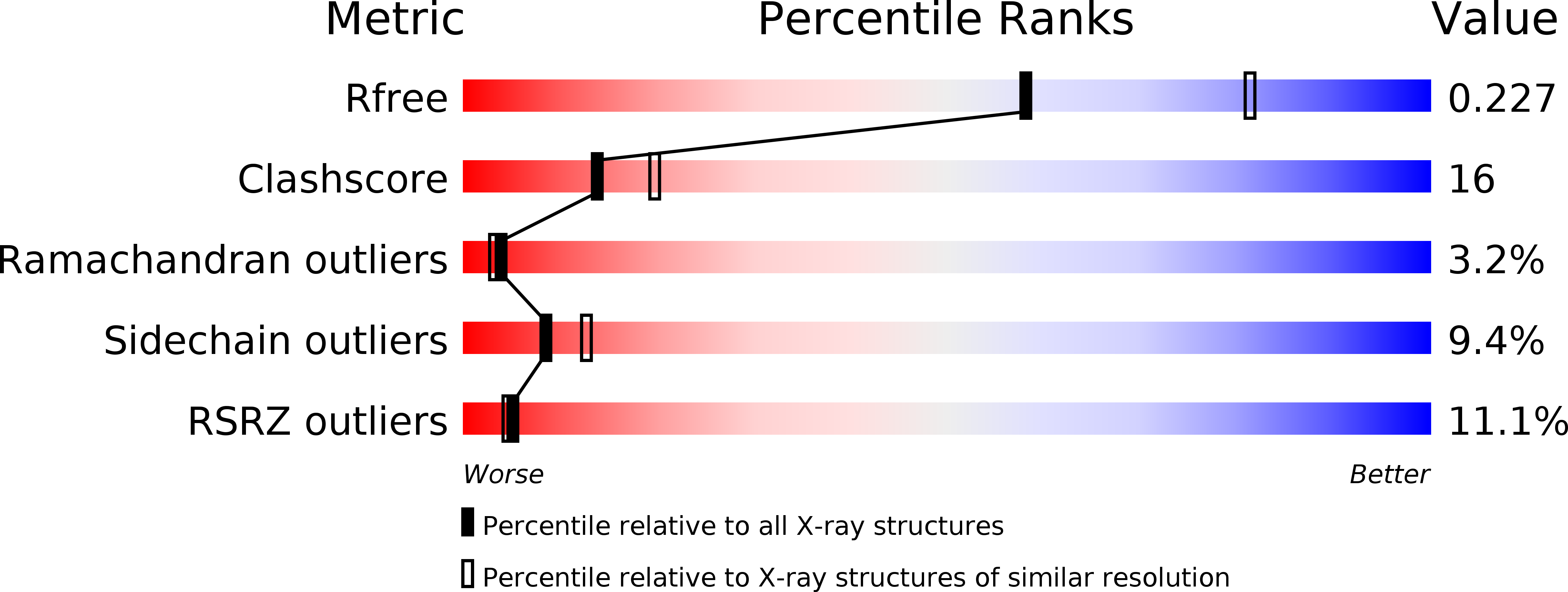
3V9R - PubMed Abstract:
Fanconi anemia (FA) is a chromosomal instability disorder associated with deficiencies in the Fanconi anemia complementation group (FANC) network. A complex consisting of FANCM-associated histone-fold proteins 1 and 2 (MHF1 and MHF2) has been shown to act cooperatively with FANCM in DNA damage repair in the FA pathway. Here we report the structure of Saccharomyces cerevisiae MHF complex in which MHF1 and MHF2 assume a typical histone fold, and the complex has a heterotetrameric architecture similar to that of the histones (H3-H4)₂ heterotetramer. Loop L2 of MHF1 is probably involved in DNA binding, and loop L3 and helices α2 and α3 of one MHF1 subunit interact with those of the other to form two heterotetramer interfaces. Further genetic data demonstrate that the heterotetramer assembly is essential for the function of the complex in DNA repair. These results provide, to the best of our knowledge, new mechanistic insights into the function of the MHF complex.
Organizational Affiliation:
State Key Laboratory of Molecular Biology and Research Center for Structural Biology, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, 320 Yue Yang Road, Shanghai 200031, China.